publications
publications by categories in reversed chronological order.
2025
-
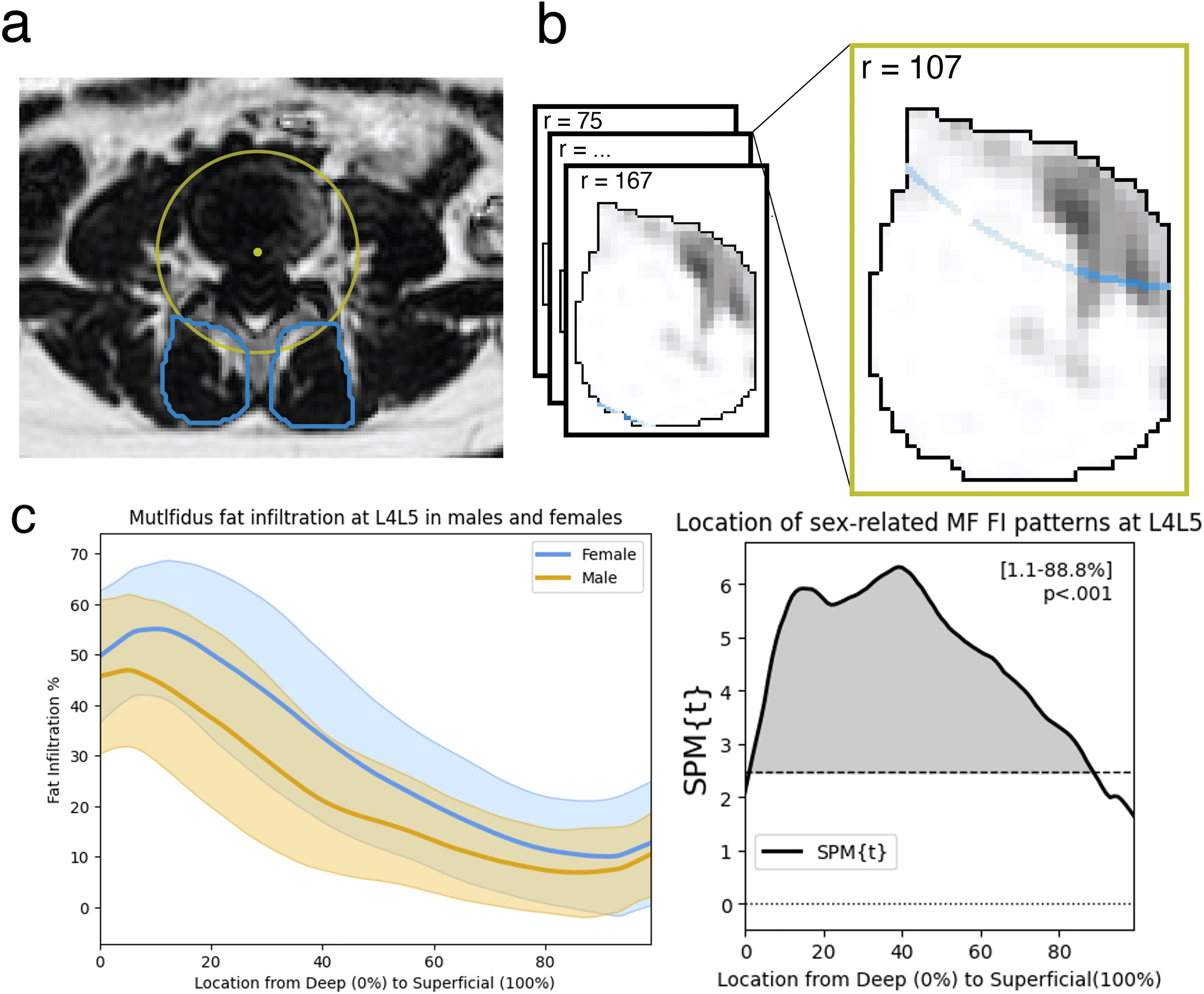 Spatial patterns of fat within the deep multifidus as a biomarker for chronic low back painKarim Khattab, Lucas K. Dziesinski, Jessica Ornowski, Jiamin Zhou, Noah B. Bonnheim, Rebecca Crawford, Aaron Scheffler, Aaron J. Fields, Conor W. O’Neill, Jeffrey C. Lotz, and Jeannie F. BaileyThe Spine Journal, Jul 2025
Spatial patterns of fat within the deep multifidus as a biomarker for chronic low back painKarim Khattab, Lucas K. Dziesinski, Jessica Ornowski, Jiamin Zhou, Noah B. Bonnheim, Rebecca Crawford, Aaron Scheffler, Aaron J. Fields, Conor W. O’Neill, Jeffrey C. Lotz, and Jeannie F. BaileyThe Spine Journal, Jul 2025Background Context: Chronic low back pain (cLBP) patients often have elevated fat infiltration (FI) in the multifidus (MF), but it is unclear how this relates to pain and degenerative spine features. Most prior work assess MF degeneration as the average whole-muscle fat content even though deep and superficial fascicles of the MF have different structural and functional characteristics. Assessing the spatial distribution of MF FI may provide regional context for causal mechanisms which may have distinct regional presentations within the muscle. Purpose: This study assesses spatial patterns of MF FI at each lumbar level to identify regional differences associated with cLBP symptoms and degenerative spine features. Study Design: This is an observational cross-sectional study. Patient Sample: Our study sample consisted of 230 cLBP patients from the BACPAC comeBack cohort who reported low back pain that has persisted for the past three months. Outcome Measures: Measures included multifidus fat-maps – a fat infiltration curve showing the spatial distribution of fat moving radially through the multifidus – created at each lumbar level (L1L2-L5S1). Other measures included average fat fraction in the deepest 15% of the MF (deep15 FI%), average whole-muscle fat fraction (overall FI%) and the Pain, Enjoyment of Life, and General Activity (PEG) survey score. Methods: We collected 3T MRI and used advanced sequences (IDEAL) to map the spatial distribution of MF FI at each lumbar level. We used statistical parametric mapping to identify spatial patterns of fat in the MF associated with age, sex, and BMI. Then, we tested for differences in spatial patterns of MF FI associated with pain and adjacent disc degeneration. Next, we calculated the fat fraction in a region of interest in the deepest 15% of the MF (deep15 FI%) and used linear mixed effects modeling to compare ho w age, sex, BMI, pain, and degenerative spine features associate with the deep15 FI% and the overall FI% separately. Lastly, we used linear regression models of PEG to compare FI measures as predictors for pain. Results: Elevated FI associated with PEG and adjacent disc degeneration only within the deepest 10% (p<.05) and deepest 25% (p<.01) of the MF respectively at the lower lumbar levels. Associations between demographic factors and FI were not specific to the deep MF. Older age and female sex associated with elevated FI throughout the muscle (p<.001) while higher BMI associated with elevated FI in only the superficial 60% of the MF (p<.001). Lastly, higher mean deep15 FI% but not mean overall FI% at the lower lumbar levels associated with higher PEG (p=.023). Conclusions: Fat infiltration in deep regions of the MF at L4L5 and L5S1 is more strongly associated with pain and adjacent disc degeneration and less associated with age, sex, and BMI than overall FI%. We identify regional differences in MF FI related to pain which improves our understanding of cLBP-related multifidus degeneration and provides additional context for possible causal mechanisms of FI. Further, the “deep15” FI% is a novel MF FI measure, and possible biomarker, more strongly associated with cLBP than summary measures.
@article{Khattab_Dziesinski_Ornowski_Zhou_Bonnheim_Crawford_Scheffler_Fields_O'Neill_Lotz_2025, title = {Spatial patterns of fat within the deep multifidus as a biomarker for chronic low back pain}, volume = {0}, issn = {1529-9430, 1878-1632}, url = {https://www.thespinejournalonline.com/article/S1529-9430(25)00341-9/fulltext}, doi = {10.1016/j.spinee.2025.07.022}, number = {0}, journal = {The Spine Journal}, publisher = {Elsevier}, author = {Khattab, Karim and Dziesinski, Lucas K. and Ornowski, Jessica and Zhou, Jiamin and Bonnheim, Noah B. and Crawford, Rebecca and Scheffler, Aaron and Fields, Aaron J. and O'Neill, Conor W. and Lotz, Jeffrey C. and Bailey, Jeannie F.}, year = {2025}, month = jul, language = {English} } -
 Artificial intelligence in musculoskeletal applications: a primer for radiologistsMichelle W. Tong, Jiamin Zhou, Zehra Akkaya, Sharmila Majumdar, Rupsa Bhattacharjee, Michelle W. Tong, Jiamin Zhou, Zehra Akkaya, Sharmila Majumdar, and Rupsa BhattacharjeeDiagnostic and Interventional Radiology, Mar 2025
Artificial intelligence in musculoskeletal applications: a primer for radiologistsMichelle W. Tong, Jiamin Zhou, Zehra Akkaya, Sharmila Majumdar, Rupsa Bhattacharjee, Michelle W. Tong, Jiamin Zhou, Zehra Akkaya, Sharmila Majumdar, and Rupsa BhattacharjeeDiagnostic and Interventional Radiology, Mar 2025As an umbrella term, artificial intelligence (AI) covers machine learning and deep learning. This review aimed to elaborate on these terms to act as a primer for radiologists to learn more about the algorithms commonly used in musculoskeletal radiology. It also aimed to familiarize them with the common practices and issues in the use of AI in this domain.
@article{Tong_Zhou_Akkaya_Majumdar_Bhattacharjee_Tong_Zhou_Akkaya_Majumdar_Bhattacharjee_2025, title = {Artificial intelligence in musculoskeletal applications: a primer for radiologists}, issn = {1305-3612}, url = {https://dirjournal.org/articles/artificial-intelligence-in-musculoskeletal-applications-a-primer-for-radiologists/dir.2024.242830}, doi = {10.4274/dir.2024.242830}, journal = {Diagnostic and Interventional Radiology}, publisher = {Diagnostic and Interventional Radiology}, author = {Tong, Michelle W. and Zhou, Jiamin and Akkaya, Zehra and Majumdar, Sharmila and Bhattacharjee, Rupsa and Tong, Michelle W. and Zhou, Jiamin and Akkaya, Zehra and Majumdar, Sharmila and Bhattacharjee, Rupsa}, year = {2025}, month = mar }
2023
-
 ISSLS Prize in Bioengineering Science 2023: Age- and sex-related differences in lumbar intervertebral disc degeneration between patients with chronic low back pain and asymptomatic controlsNoah B. Bonnheim, Ann A. Lazar, Anika Kumar, Zehra Akkaya, Jiamin Zhou, Xiaojie Guo, Conor O’Neill, Thomas M. Link, Jeffrey C. Lotz, Roland Krug, and Aaron J. FieldsEuropean Spine Journal, May 2023
ISSLS Prize in Bioengineering Science 2023: Age- and sex-related differences in lumbar intervertebral disc degeneration between patients with chronic low back pain and asymptomatic controlsNoah B. Bonnheim, Ann A. Lazar, Anika Kumar, Zehra Akkaya, Jiamin Zhou, Xiaojie Guo, Conor O’Neill, Thomas M. Link, Jeffrey C. Lotz, Roland Krug, and Aaron J. FieldsEuropean Spine Journal, May 2023Clinical management of disc degeneration in patients with chronic low back pain (cLBP) is hampered by the challenge of distinguishing pathologic changes relating to pain from physiologic changes related to aging. The goal of this study was to use imaging biomarkers of disc biochemical composition to distinguish degenerative changes associated with cLBP from normal aging.
@article{Bonnheim_Lazar_Kumar_Akkaya_Zhou_Guo_O'Neill_Link_Lotz_Krug_2023, title = {ISSLS Prize in Bioengineering Science 2023: Age- and sex-related differences in lumbar intervertebral disc degeneration between patients with chronic low back pain and asymptomatic controls}, volume = {32}, issn = {1432-0932}, doi = {10.1007/s00586-023-07542-6}, number = {5}, journal = {European Spine Journal}, author = {Bonnheim, Noah B. and Lazar, Ann A. and Kumar, Anika and Akkaya, Zehra and Zhou, Jiamin and Guo, Xiaojie and O'Neill, Conor and Link, Thomas M. and Lotz, Jeffrey C. and Krug, Roland and Fields, Aaron J.}, year = {2023}, month = may, pages = {1517–1524}, language = {en} } -
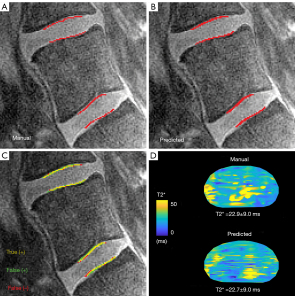 Deep-learning-based biomarker of spinal cartilage endplate health using ultra-short echo time magnetic resonance imagingNoah B. Bonnheim, Linshanshan Wang, Ann A. Lazar, Ravi Chachad, Jiamin Zhou, Xiaojie Guo, Conor O’Neill, Joel Castellanos, Jiang Du, Hyungseok Jang, Roland Krug, and Aaron J. FieldsQuantitative Imaging in Medicine and Surgery, May 2023
Deep-learning-based biomarker of spinal cartilage endplate health using ultra-short echo time magnetic resonance imagingNoah B. Bonnheim, Linshanshan Wang, Ann A. Lazar, Ravi Chachad, Jiamin Zhou, Xiaojie Guo, Conor O’Neill, Joel Castellanos, Jiang Du, Hyungseok Jang, Roland Krug, and Aaron J. FieldsQuantitative Imaging in Medicine and Surgery, May 2023Background: T2* relaxation times in the spinal cartilage endplate (CEP) measured using ultra-short echo time magnetic resonance imaging (UTE MRI) reflect aspects of biochemical composition that influence the CEP’s permeability to nutrients. Deficits in CEP composition measured using T2* biomarkers from UTE MRI are associated with more severe intervertebral disc degeneration in patients with chronic low back pain (cLBP). The goal of this study was to develop an objective, accurate, and efficient deep-learning-based method for calculating biomarkers of CEP health using UTE images. Methods: Multi-echo UTE MRI of the lumbar spine was acquired from a prospectively enrolled cross-sectional and consecutive cohort of 83 subjects spanning a wide range of ages and cLBP-related conditions. CEPs from the L4-S1 levels were manually segmented on 6,972 UTE images and used to train neural networks utilizing the u-net architecture. CEP segmentations and mean CEP T2* values derived from manually- and model-generated segmentations were compared using Dice scores, sensitivity, specificity, Bland-Altman, and receiver-operator characteristic (ROC) analysis. Signal-to-noise (SNR) and contrast-to-noise (CNR) ratios were calculated and related to model performance. Results: Compared with manual CEP segmentations, model-generated segmentations achieved sensitives of 0.80–0.91, specificities of 0.99, Dice scores of 0.77–0.85, area under the receiver-operating characteristic curve values of 0.99, and precision-recall (PR) AUC values of 0.56–0.77, depending on spinal level and sagittal image position. Mean CEP T2* values and principal CEP angles derived from the model-predicted segmentations had low bias in an unseen test dataset (T2* bias =0.33±2.37 ms, angle bias =0.36±2.65°). To simulate a hypothetical clinical scenario, the predicted segmentations were used to stratify CEPs into high, medium, and low T2* groups. Group predictions had diagnostic sensitivities of 0.77–0.86 and specificities of 0.86–0.95. Model performance was positively associated with image SNR and CNR. Conclusions: The trained deep learning models enable accurate, automated CEP segmentations and T2* biomarker computations that are statistically similar to those from manual segmentations. These models address limitations with inefficiency and subjectivity associated with manual methods. Such techniques could be used to elucidate the role of CEP composition in disc degeneration etiology and guide emerging therapies for cLBP.
@article{Bonnheim_Wang_Lazar_Chachad_Zhou_Guo_O'Neill_Castellanos_Du_Jang_2023, title = {Deep-learning-based biomarker of spinal cartilage endplate health using ultra-short echo time magnetic resonance imaging}, volume = {13}, issn = {2223-4306, 2223-4292}, doi = {10.21037/qims-22-729}, number = {5}, journal = {Quantitative Imaging in Medicine and Surgery}, publisher = {AME Publishing Company}, author = {Bonnheim, Noah B. and Wang, Linshanshan and Lazar, Ann A. and Chachad, Ravi and Zhou, Jiamin and Guo, Xiaojie and O'Neill, Conor and Castellanos, Joel and Du, Jiang and Jang, Hyungseok and Krug, Roland and Fields, Aaron J.}, year = {2023}, month = may, pages = {2807821–2802821}, language = {en} }
2022
-
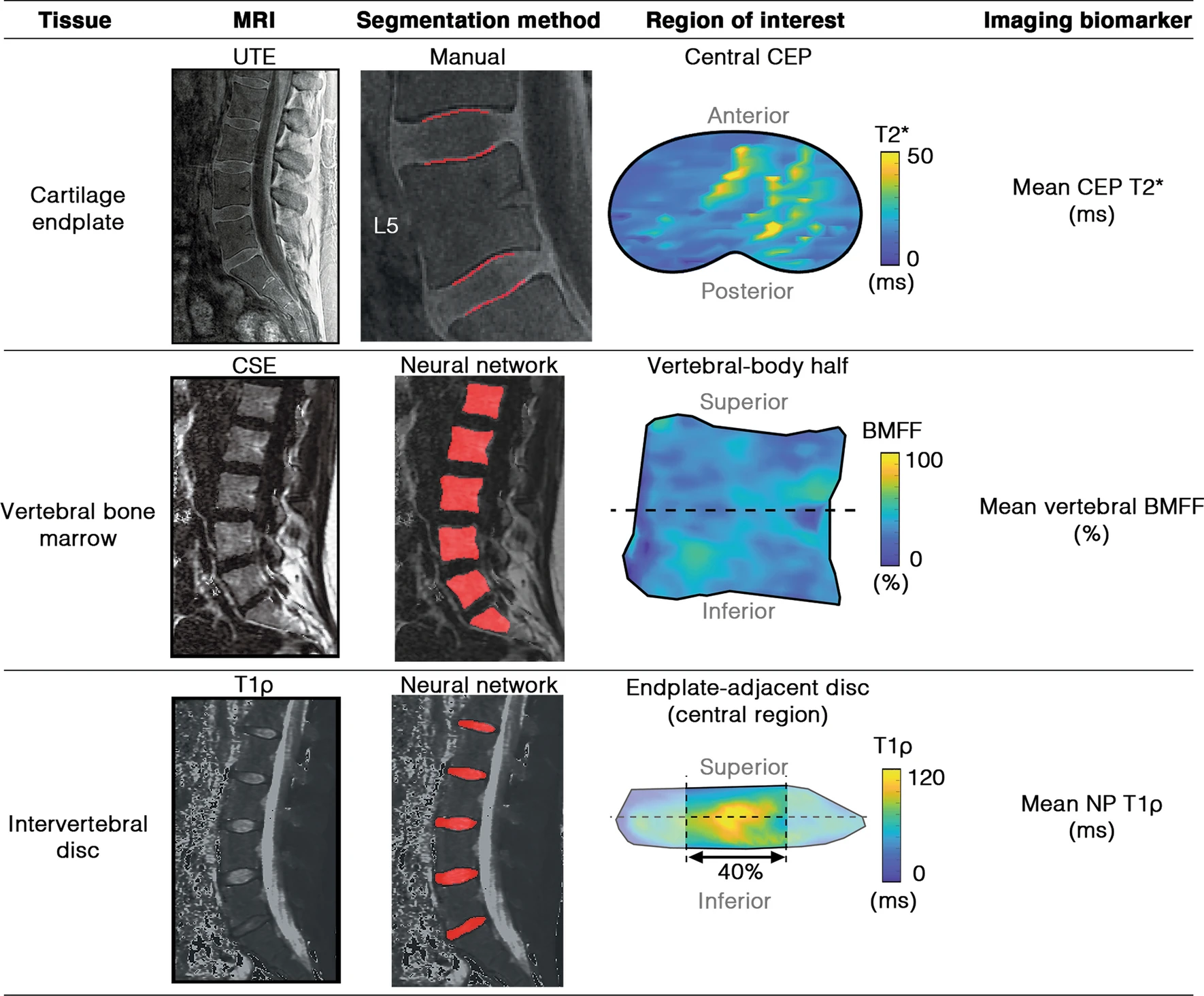 The contributions of cartilage endplate composition and vertebral bone marrow fat to intervertebral disc degeneration in patients with chronic low back painNoah B. Bonnheim, Linshanshan Wang, Ann A. Lazar, Jiamin Zhou, Ravi Chachad, Nico Sollmann, Xiaojie Guo, Claudia Iriondo, Conor O’Neill, Jeffrey C. Lotz, Thomas M. Link, Roland Krug, and Aaron J. FieldsEuropean Spine Journal, Jul 2022
The contributions of cartilage endplate composition and vertebral bone marrow fat to intervertebral disc degeneration in patients with chronic low back painNoah B. Bonnheim, Linshanshan Wang, Ann A. Lazar, Jiamin Zhou, Ravi Chachad, Nico Sollmann, Xiaojie Guo, Claudia Iriondo, Conor O’Neill, Jeffrey C. Lotz, Thomas M. Link, Roland Krug, and Aaron J. FieldsEuropean Spine Journal, Jul 2022The composition of the subchondral bone marrow and cartilage endplate (CEP) could affect intervertebral disc health by influencing vertebral perfusion and nutrient diffusion. However, the relative contributions of these factors to disc degeneration in patients with chronic low back pain (cLBP) have not been quantified. The goal of this study was to use compositional biomarkers derived from quantitative MRI to establish how CEP composition (surrogate for permeability) and vertebral bone marrow fat fraction (BMFF, surrogate for perfusion) relate to disc degeneration.
@article{Bonnheim_Wang_Lazar_Zhou_Chachad_Sollmann_Guo_Iriondo_O'Neill_Lotz_2022, title = {The contributions of cartilage endplate composition and vertebral bone marrow fat to intervertebral disc degeneration in patients with chronic low back pain}, volume = {31}, issn = {1432-0932}, doi = {10.1007/s00586-022-07206-x}, number = {7}, journal = {European Spine Journal}, author = {Bonnheim, Noah B. and Wang, Linshanshan and Lazar, Ann A. and Zhou, Jiamin and Chachad, Ravi and Sollmann, Nico and Guo, Xiaojie and Iriondo, Claudia and O'Neill, Conor and Lotz, Jeffrey C. and Link, Thomas M. and Krug, Roland and Fields, Aaron J.}, year = {2022}, month = jul, pages = {1866–1872}, language = {en} } -
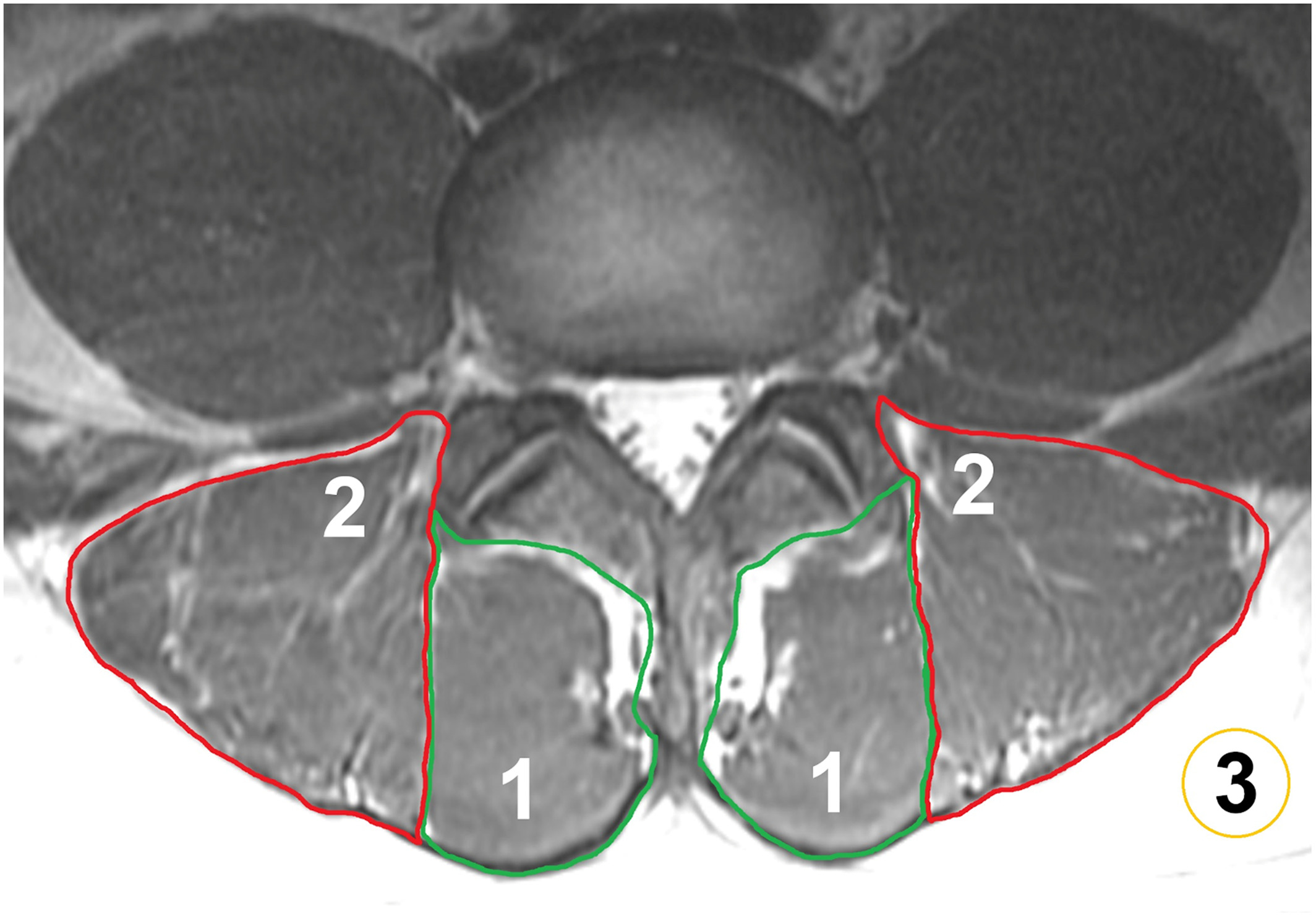 Paraspinal Muscle in Chronic Low Back Pain: Comparison Between Standard Parameters and Chemical Shift Encoding-Based Water–Fat MRINico Sollmann, Noah B. Bonnheim, Gabby B. Joseph, Ravi Chachad, Jiamin Zhou, Zehra Akkaya, Amir M. Pirmoazen, Jeannie F. Bailey, Xiaojie Guo, Ann A. Lazar, Thomas M. Link, Aaron J. Fields, and Roland KrugJournal of Magnetic Resonance Imaging, Jul 2022
Paraspinal Muscle in Chronic Low Back Pain: Comparison Between Standard Parameters and Chemical Shift Encoding-Based Water–Fat MRINico Sollmann, Noah B. Bonnheim, Gabby B. Joseph, Ravi Chachad, Jiamin Zhou, Zehra Akkaya, Amir M. Pirmoazen, Jeannie F. Bailey, Xiaojie Guo, Ann A. Lazar, Thomas M. Link, Aaron J. Fields, and Roland KrugJournal of Magnetic Resonance Imaging, Jul 2022Background: Paraspinal musculature (PSM) is increasingly recognized as a contributor to low back pain (LBP), but with conventional MRI sequences, assessment is limited. Chemical shift encoding-based water–fat MRI (CSE-MRI) enables the measurement of PSM fat fraction (FF), which may assist investigations of chronic LBP. Purpose: To investigate associations between PSM parameters from conventional MRI and CSE-MRI and between PSM parameters and pain. Study Type: Prospective, cross-sectional. Population: Eighty-four adults with chronic LBP (44.6 ± 13.4 years; 48 males). Field Strength/Sequence: 3-T, T1-weighted fast spin-echo and iterative decomposition of water and fat with echo asymmetry and least squares estimation sequences. Assessment: T1-weighted images for Goutallier classification (GC), muscle volume, lumbar indentation value, and muscle-fat index, CSE-MRI for FF extraction (L1/2–L5/S1). Pain was self-reported using a visual analogue scale (VAS). Intra- and/or interreader agreement was assessed for MRI-derived parameters. Statistical Tests: Mixed-effects and linear regression models to 1) assess relationships between PSM parameters (entire cohort and subgroup with GC grades 0 and 1; statistical significance α = 0.0025) and 2) evaluate associations of PSM parameters with pain (α = 0.05). Intraclass correlation coefficients (ICCs) for intra- and/or interreader agreement. Results: The FF showed excellent intra- and interreader agreement (ICC range: 0.97–0.99) and was significantly associated with GC at all spinal levels. Subgroup analysis suggested that early/subtle changes in PSM are detectable with FF but not with GC, given the absence of significant associations between FF and GC (P-value range: 0.036 at L5/S1 to 0.784 at L2/L3). Averaged over all spinal levels, FF and GC were significantly associated with VAS scores. Data Conclusion: In the absence of FF, GC may be the best surrogate for PSM quality. Given the ability of CSE-MRI to detect muscle alterations at early stages of PSM degeneration, this technique may have potential for further investigations of the role of PSM in chronic LBP. Level of Evidence: 2 Technical Efficacy Stage: 2
@article{Sollmann_Bonnheim_Joseph_Chachad_Zhou_Akkaya_Pirmoazen_Bailey_Guo_Lazar_2022, title = {Paraspinal Muscle in Chronic Low Back Pain: Comparison Between Standard Parameters and Chemical Shift Encoding-Based Water–Fat MRI}, volume = {56}, rights = {© 2022 The Authors. Journal of Magnetic Resonance Imaging published by Wiley Periodicals LLC on behalf of International Society for Magnetic Resonance in Medicine.}, issn = {1522-2586}, doi = {10.1002/jmri.28145}, number = {5}, journal = {Journal of Magnetic Resonance Imaging}, author = {Sollmann, Nico and Bonnheim, Noah B. and Joseph, Gabby B. and Chachad, Ravi and Zhou, Jiamin and Akkaya, Zehra and Pirmoazen, Amir M. and Bailey, Jeannie F. and Guo, Xiaojie and Lazar, Ann A. and Link, Thomas M. and Fields, Aaron J. and Krug, Roland}, year = {2022}, pages = {1600–1608}, language = {en} }
2020
-
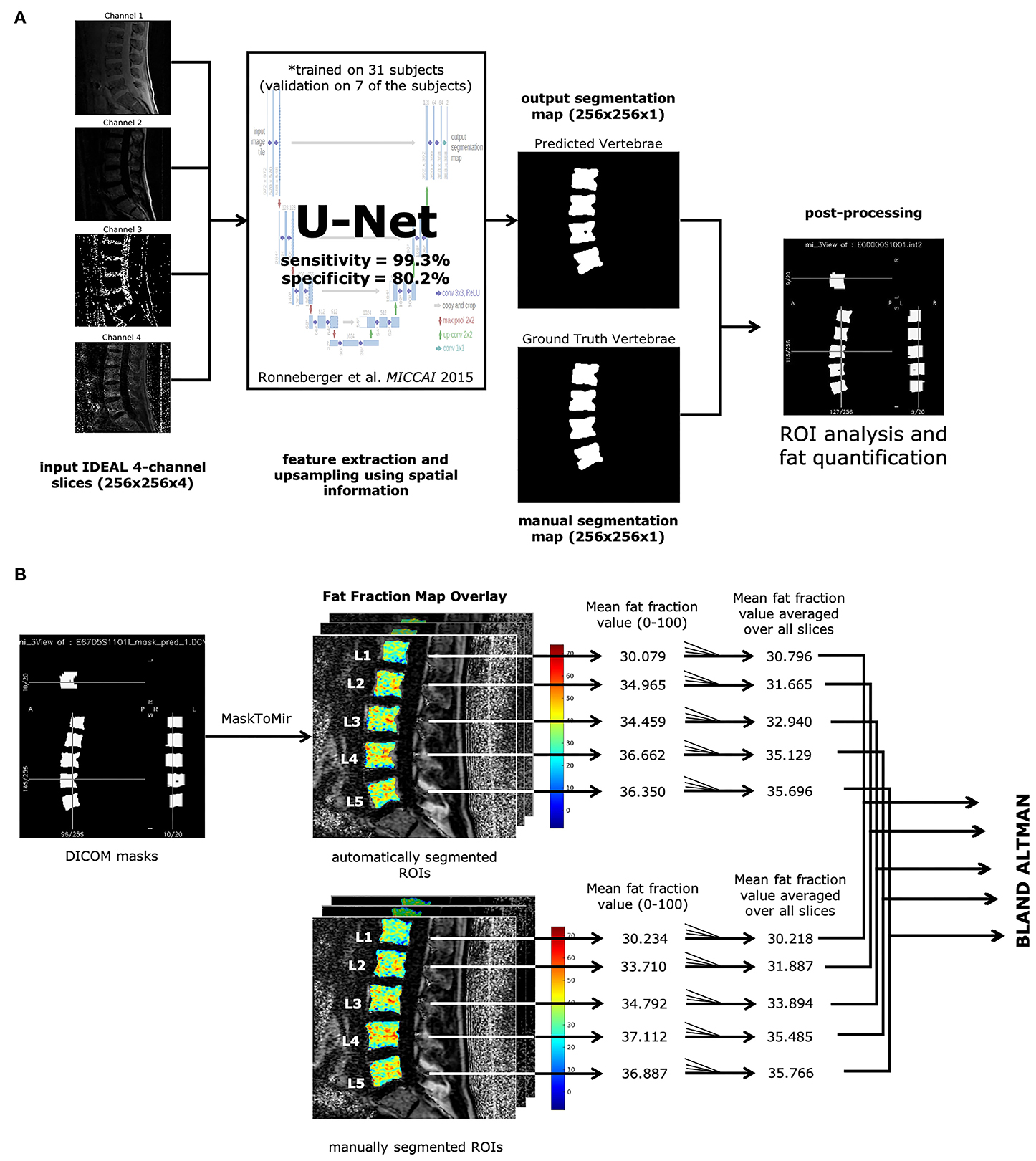 Automatic Vertebral Body Segmentation Based on Deep Learning of Dixon Images for Bone Marrow Fat Fraction QuantificationJiamin Zhou, Pablo F. Damasceno, Ravi Chachad, Justin R. Cheung, Alexander Ballatori, Jeffrey C. Lotz, Ann A. Lazar, Thomas M. Link, Aaron J. Fields, and Roland KrugFrontiers in Endocrinology, Sep 2020
Automatic Vertebral Body Segmentation Based on Deep Learning of Dixon Images for Bone Marrow Fat Fraction QuantificationJiamin Zhou, Pablo F. Damasceno, Ravi Chachad, Justin R. Cheung, Alexander Ballatori, Jeffrey C. Lotz, Ann A. Lazar, Thomas M. Link, Aaron J. Fields, and Roland KrugFrontiers in Endocrinology, Sep 2020Background: Bone marrow fat (BMF) fraction quantification in vertebral bodies is used as a novel imaging biomarker to assess and characterize chronic lower back pain. However, manual segmentation of vertebral bodies is time consuming and laborious. Purpose: (1) Develop a deep learning pipeline for segmentation of vertebral bodies using quantitative water-fat MRI. (2) Compare BMF measurements between manual and automatic segmentation methods to assess performance. Materials and Methods: In this retrospective study, MR images using a 3D spoiled gradient-recalled echo (SPGR) sequence with Iterative Decomposition of water and fat with Echo Asymmetry and Least-squares estimation (IDEAL) reconstruction algorithm were obtained in 57 subjects (28 women, 29 men, mean age, 47.2 ± 12.6 years). An artificial network was trained for 100 epochs on a total of 165 lumbar vertebrae manually segmented from 31 subjects. Performance was assessed by analyzing the receiver operating characteristic curve, precision-recall, F1 scores, specificity, sensitivity, and similarity metrics. Bland-Altman analysis was used to assess performance of BMF fraction quantification using the predicted segmentations. Results: The deep learning segmentation method achieved an AUC of 0.92 (CI 95%: 0.9186, 0.9195) on a testing dataset (n = 24 subjects) on classification of pixels as vertebrae. A sensitivity of 0.99 and specificity of 0.80 were achieved for a testing dataset, and a mean Dice similarity coefficient of 0.849 ± 0.091. Comparing manual and automatic segmentations on fat fraction maps of lumbar vertebrae (n = 124 vertebral bodies) using Bland-Altman analysis resulted in a bias of only −0.605% (CI 95% = −0.847 to −0.363%) and agreement limits of −3.275% and +2.065%. Automatic segmentation was also feasible in 16 ± 1 s. Conclusion: Our results have demonstrated the feasibility of automated segmentation of vertebral bodies using deep learning models on water-fat MR (Dixon) images to define vertebral regions of interest with high specificity. These regions of interest can then be used to quantify BMF with comparable results as manual segmentation, providing a framework for completely automated investigation of vertebral changes in CLBP.
@article{Zhou_Damasceno_Chachad_Cheung_Ballatori_Lotz_Lazar_Link_Fields_Krug_2020, title = {Automatic Vertebral Body Segmentation Based on Deep Learning of Dixon Images for Bone Marrow Fat Fraction Quantification}, volume = {11}, issn = {1664-2392}, url = {https://www.frontiersin.orghttps://www.frontiersin.org/journals/endocrinology/articles/10.3389/fendo.2020.00612/full}, doi = {10.3389/fendo.2020.00612}, journal = {Frontiers in Endocrinology}, publisher = {Frontiers}, author = {Zhou, Jiamin and Damasceno, Pablo F. and Chachad, Ravi and Cheung, Justin R. and Ballatori, Alexander and Lotz, Jeffrey C. and Lazar, Ann A. and Link, Thomas M. and Fields, Aaron J. and Krug, Roland}, year = {2020}, month = sep, language = {English} }
2019
-
 Establishing Cell Lines Overexpressing DR3 to Assess the Apoptotic Response to Anti-mitotic TherapeuticsXin Wang, Jiamin Zhou, Chen Qi, and Gelin WangJournal of Visualized Experiments (JoVE), Jan 2019
Establishing Cell Lines Overexpressing DR3 to Assess the Apoptotic Response to Anti-mitotic TherapeuticsXin Wang, Jiamin Zhou, Chen Qi, and Gelin WangJournal of Visualized Experiments (JoVE), Jan 2019Studying the function of a gene of interest can be achieved by manipulating its level of expression, such as decreasing its expression with knockdown cell lines or increasing its expression with overexpression cell lines. Transient and stable transfection are two methods that are often used for exogenous gene expression. Transient transfection is only useful for short-term expression, whereas stable transfection allows exogenous genes to be integrated into the host cell genome where it will be continuously expressed. As a result, stable transfection is usually employed for research into long-term genetic regulation. Here we describe a simple protocol to generate a stable cell line overexpressing tagged death receptor 3 (DR3) to explore DR3 function. We picked single clones after a retroviral infection in order to maintain the homogeneity and purity of the stable cell lines. The stable cell lines generated using this protocol render DR3-deficient HT29 cells sensitive to antimitotic drugs, thus reconstituting the apoptotic response in HT29 cells. Moreover, the FLAG tag on DR3 compensates for the unavailability of good DR3 antibody and facilitates the biochemical study of the molecular mechanism by which antimitotic agents induce apoptosis.
@article{Wang_Zhou_Qi_Wang_2019, title = {Establishing Cell Lines Overexpressing DR3 to Assess the Apoptotic Response to Anti-mitotic Therapeutics}, issn = {1940-087X}, doi = {10.3791/58705}, number = {143}, journal = {Journal of Visualized Experiments (JoVE)}, author = {Wang, Xin and Zhou, Jiamin and Qi, Chen and Wang, Gelin}, year = {2019}, month = jan, pages = {e58705}, language = {en} }
2018
-
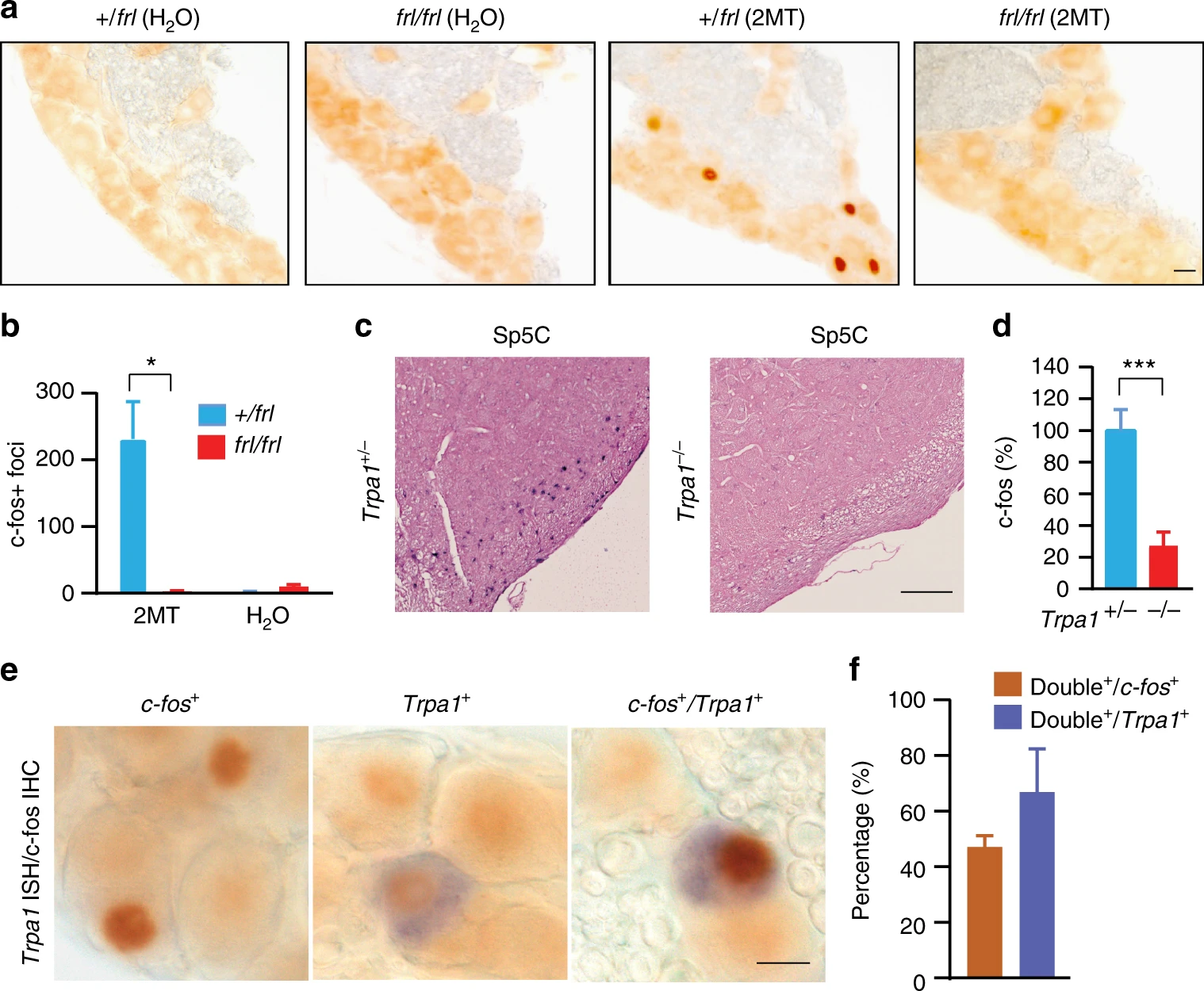 Large-scale forward genetics screening identifies Trpa1 as a chemosensor for predator odor-evoked innate fear behaviorsYibing Wang, Liqin Cao, Chia-Ying Lee, Tomohiko Matsuo, Kejia Wu, Greg Asher, Lijun Tang, Tsuyoshi Saitoh, Jamie Russell, Daniela Klewe-Nebenius, Li Wang, Shingo Soya, Emi Hasegawa, Yoan Chérasse, Jiamin Zhou, Yuwenbin Li, Tao Wang, Xiaowei Zhan, Chika Miyoshi, Yoko Irukayama, Jie Cao, Julian P. Meeks, Laurent Gautron, Zhiqiang Wang, Katsuyasu Sakurai, Hiromasa Funato, Takeshi Sakurai, Masashi Yanagisawa, Hiroshi Nagase, Reiko Kobayakawa, Ko Kobayakawa, Bruce Beutler, and Qinghua LiuNature Communications, May 2018
Large-scale forward genetics screening identifies Trpa1 as a chemosensor for predator odor-evoked innate fear behaviorsYibing Wang, Liqin Cao, Chia-Ying Lee, Tomohiko Matsuo, Kejia Wu, Greg Asher, Lijun Tang, Tsuyoshi Saitoh, Jamie Russell, Daniela Klewe-Nebenius, Li Wang, Shingo Soya, Emi Hasegawa, Yoan Chérasse, Jiamin Zhou, Yuwenbin Li, Tao Wang, Xiaowei Zhan, Chika Miyoshi, Yoko Irukayama, Jie Cao, Julian P. Meeks, Laurent Gautron, Zhiqiang Wang, Katsuyasu Sakurai, Hiromasa Funato, Takeshi Sakurai, Masashi Yanagisawa, Hiroshi Nagase, Reiko Kobayakawa, Ko Kobayakawa, Bruce Beutler, and Qinghua LiuNature Communications, May 2018Innate behaviors are genetically encoded, but their underlying molecular mechanisms remain largely unknown. Predator odor 2,4,5-trimethyl-3-thiazoline (TMT) and its potent analog 2-methyl-2-thiazoline (2MT) are believed to activate specific odorant receptors to elicit innate fear/defensive behaviors in naive mice. Here, we conduct a large-scale recessive genetics screen of ethylnitrosourea (ENU)-mutagenized mice. We find that loss of Trpa1, a pungency/irritancy receptor, diminishes TMT/2MT and snake skin-evoked innate fear/defensive responses. Accordingly, Trpa1−/−mice fail to effectively activate known fear/stress brain centers upon 2MT exposure, despite their apparent ability to smell and learn to fear 2MT. Moreover, Trpa1 acts as a chemosensor for 2MT/TMT and Trpa1-expressing trigeminal ganglion neurons contribute critically to 2MT-evoked freezing. Our results indicate that Trpa1-mediated nociception plays a crucial role in predator odor-evoked innate fear/defensive behaviors. The work establishes the first forward genetics screen to uncover the molecular mechanism of innate fear, a basic emotion and evolutionarily conserved survival mechanism.
@article{Wang_Cao_Lee_Matsuo_Wu_Asher_Tang_Saitoh_Russell_Klewe-Nebenius_2018, title = {Large-scale forward genetics screening identifies Trpa1 as a chemosensor for predator odor-evoked innate fear behaviors}, volume = {9}, rights = {2018 The Author(s)}, issn = {2041-1723}, doi = {10.1038/s41467-018-04324-3}, number = {1}, journal = {Nature Communications}, publisher = {Nature Publishing Group}, author = {Wang, Yibing and Cao, Liqin and Lee, Chia-Ying and Matsuo, Tomohiko and Wu, Kejia and Asher, Greg and Tang, Lijun and Saitoh, Tsuyoshi and Russell, Jamie and Klewe-Nebenius, Daniela and Wang, Li and Soya, Shingo and Hasegawa, Emi and Chérasse, Yoan and Zhou, Jiamin and Li, Yuwenbin and Wang, Tao and Zhan, Xiaowei and Miyoshi, Chika and Irukayama, Yoko and Cao, Jie and Meeks, Julian P. and Gautron, Laurent and Wang, Zhiqiang and Sakurai, Katsuyasu and Funato, Hiromasa and Sakurai, Takeshi and Yanagisawa, Masashi and Nagase, Hiroshi and Kobayakawa, Reiko and Kobayakawa, Ko and Beutler, Bruce and Liu, Qinghua}, year = {2018}, month = may, pages = {2041}, language = {en} }