verteseg
Automatic vertebral body segmentation on IDEAL MRI
My first first-author publication is a technical article on using a deep learning pipeline for segmentation of vertebral bodies using quantitative water-fat MRI.
If you’re interested in learning more, here is the full article.
Background
Studies have shown that increased bone marrow fat (BMF) in the vertebral bodies is linked to conditions like osteoporosis, chronic low back pain, intervertebral disc degeneration, and metabolic disorders including HIV and diabetes. These associations suggest that BMF could serve as an early biomarker for disease, offering clues about bone strength, inflammation, or metabolic shifts before more obvious symptoms or structural changes appear.
While BMF has clinical relevance, measuring it accurately isn’t straightforward. Conventional MRI sequences like T1- and T2-weighted imaging can give a general sense of fat content based on tissue brightness, but these methods are subjective and not reliable for longitudinal tracking or large-scale studies. A more precise approach is needed to quantify fat content objectively and reproducibly.
IDEAL MRI
IDEAL MRI (Iterative Decomposition of water and fat with Echo Asymmetry and Least-squares estimation) is an MRI technique that builds on Dixon imaging principles, separating water and fat signals in each voxel to create detailed fat-fraction maps
Approach
U-Net
To streamline the segmentation process, we turned to deep learning. Specifically, we used a U-Net convolutional neural network, which has become a standard tool for medical image segmentation
Training
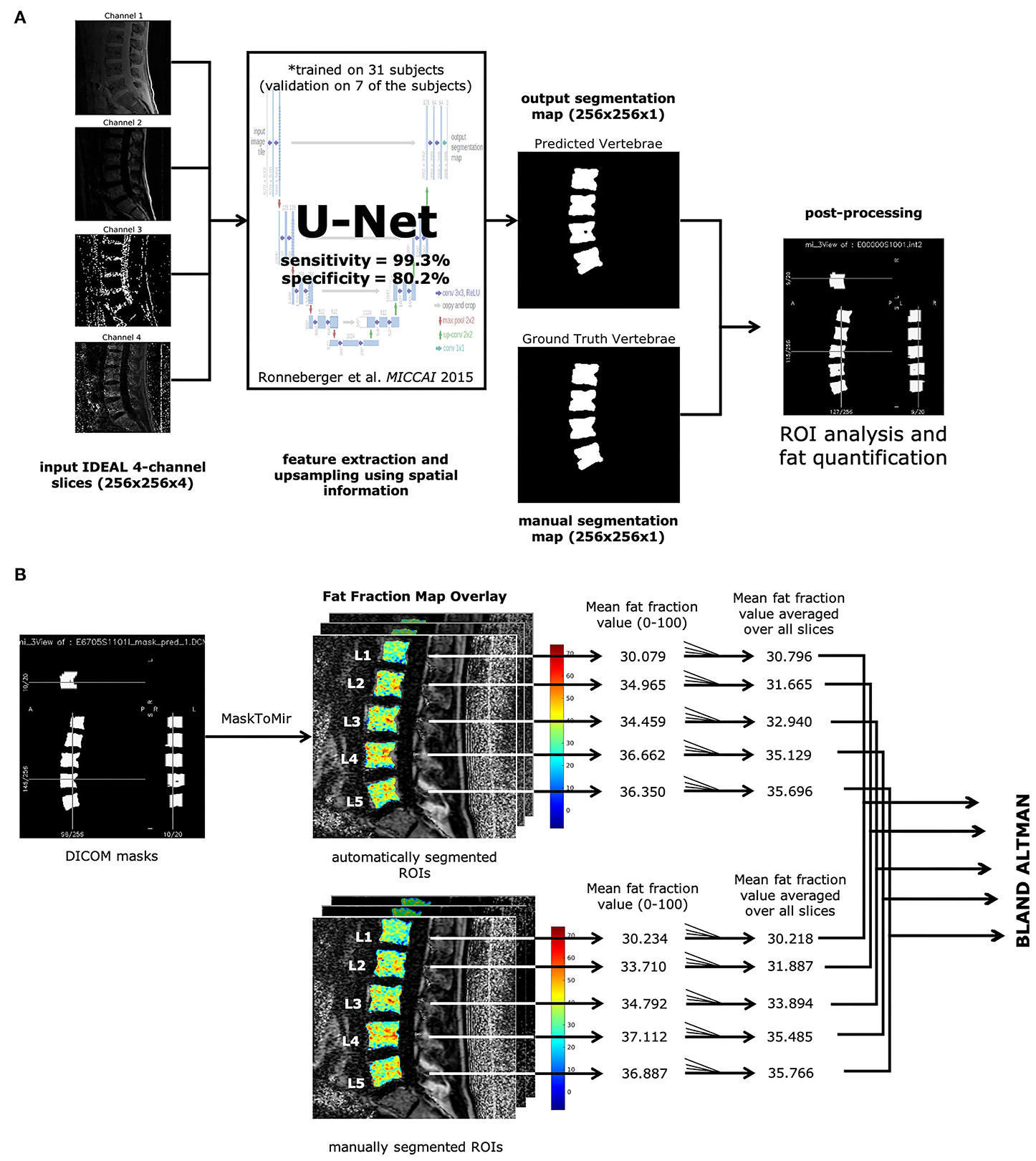
Manual segmentations of the lumbar vertebral bodies from L1-L5 were first created on IDEAL MRI to serve as training data for the U-Net. Of 57 total subjects segmented, 31 were used to train the model, while the rest were held out for validation. The goal was for the network to learn to automatically produce vertebral masks from new, unseen scans without needing further human input.
Once the U-Net model segmented the vertebral bodies, those masks were applied to the IDEAL fat-fraction maps. This allowed the pipeline to calculate BMF for each vertebral body based entirely on the automated output. In effect, the process became fully automated: starting with raw MRI images and ending with precise fat measurements, all without manual intervention at the point of use.
Results
Code can be found online.
Segmentation Accuracy
The U-Net performed well on unseen data. In testing, the model achieved an area under the ROC curve (AUC) of approximately 0.92, with a Dice similarity coefficient around 0.85, indicating strong overlap between the automated and manual vertebral masks. Sensitivity was very high (~0.99), meaning the model rarely missed vertebral regions. Specificity was slightly lower (~0.80), reflecting some inclusion of nearby tissue, but within acceptable bounds for clinical applications.
Quantifying BMF
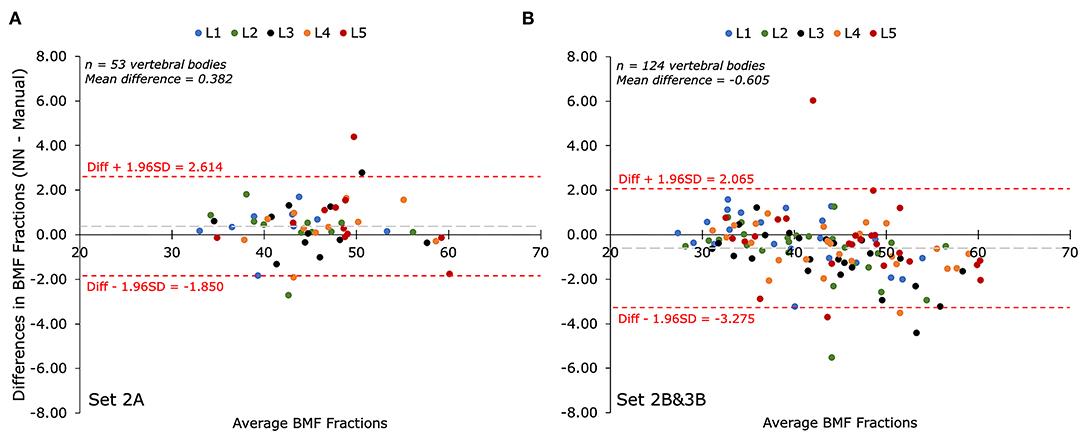
When comparing BMF values calculated by the automated segmentations to those obtained through manual segmentation, the differences were minimal. The average bias was about 0.6%, and the limits of agreement were within ±3.3%. This level of concordance suggests the automated system can reliably replicate expert-level measurement precision.
Workflow Efficienty and Reliability
Manual segmentation of the vertebrae took about 5 minutes per scan. The U-Net model completed the same task in roughly 16 seconds, which is a 92-fold improvement in speed. This efficiency could make BMF quantification feasible in settings where time and labor are limited, such as large research cohorts or busy clinical practices.
The model also demonstrated excellent repeatability. In test-retest scans, the precision error was just 1.6%, and the intraclass correlation coefficient (ICC) was a perfect 1.00. This means that repeated measurements on the same subject were virtually identical, an essential requirement for tracking changes over time.
By combining the accuracy of IDEAL MRI with the efficiency of deep learning–based segmentation, we presented a practical and scalable solution for bone marrow fat quantification. This opens the door for BMF to be used more widely, not just in specialized research studies, but potentially in routine imaging for patients with back pain, metabolic disorders, or bone health concerns.